Columbia University
Irving Medical Center
Neurological Institute
710 West 168th Street, 3rd floor
(212) 305-1818
TaubCONNECT Research Perspective:
July 2024
2: Extended Genome-Wide Association Study Employing the African Genome Resources Panel Identifies Novel Susceptibility Loci for Alzheimer's Disease in Individuals of African Ancestry
3: Adult-Onset Deactivation of Autophagy Leads to loss of Synapse Homeostasis and Cognitive Impairment, with Implications for Alzheimer Disease
 |
 |
| Vilas Menon, PhD | Gunnar Hargus, MD, PhD |
Alzheimer’s disease (AD) is the most frequent form of dementia affecting millions of people worldwide. Given the limited treatment options for AD patients and the growing demand for disease modeling platforms using human cells, induced pluripotent stem cells (iPSCs) have emerged as a powerful tool to examine AD phenotypes in neurons and glial cells differentiated from these iPSCs. This technology has been successfully applied to study tau and Aβ pathology in cultured neurons from patients with familial AD in vitro. However, adequate cell–cell interactions, synaptic function and the systematic complexity of the brain microenvironment are difficult to model in vitro, providing a strong rationale to evaluate disease-associated changes in human AD neurons in vivo, e.g. by injecting the human cells into the brains of adult mice.
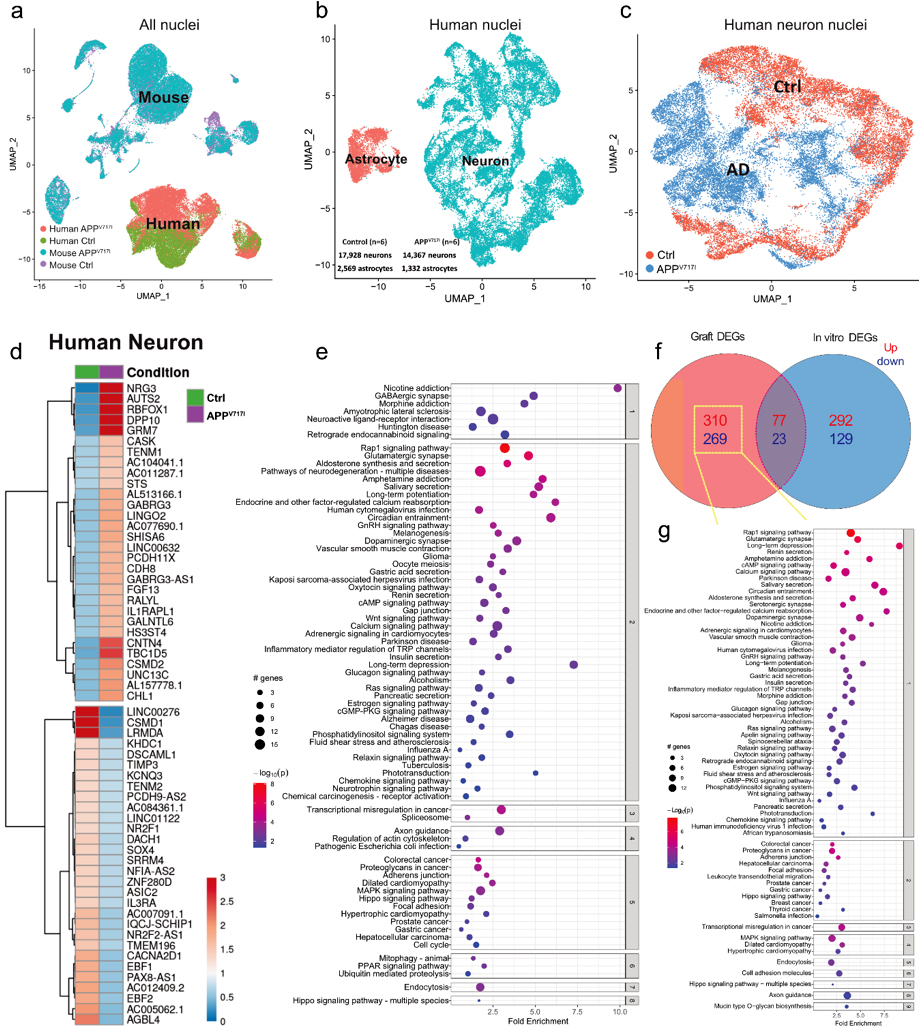
In a recent study published in Acta Neuropathologica, our groups explored pathological changes in human iPSC-derived neurons carrying the APPV717I mutation to model familial AD. APPV717I mutant iPSCs and isogenic controls were differentiated into neurons using an established protocol revealing enhanced production of hyperphosphorylated tau protein, elevated levels of Aβ42 with an increased Aβ42/Aβ40 ratio, as well as neurite outgrowth deficits in APPV717I neurons in vitro. When injected into the forebrain of immunocompromised adult mice, APPV717I and control neural cells showed robust engraftment with formation of viable neuronal grafts 2 months after transplantation. However, at 12 months post-injection, APPV717I grafts were smaller and demonstrated impaired neurite outgrowth into adjacent and remote brain regions compared to control grafts. Single-nucleus RNA-sequencing of micro-dissected grafts, performed 2 months after cell injection, identified significantly altered transcriptome signatures in APPV717I iPSC-derived neurons pointing towards dysregulated synaptic function and axon guidance. Interestingly, APPV717I neurons showed an increased expression of genes, many of which are also upregulated in postmortem cortical neurons of AD patients, including the transmembrane protein LINGO2. Downregulation of LINGO2 in cultured APPV717I neurons rescued neurite outgrowth deficits and reversed key AD-associated transcriptional changes related but not limited to synaptic function, apoptosis, and cellular senescence. These results provided important insights into transcriptional dysregulation in xenografted APPV717I neurons linked to synaptic function, and they indicated that LINGO2 may represent a potential therapeutic target in AD, which should be further explored in future studies.
|
Figure 1. Single-nucleus RNA-sequencing of APPV717I and Ctrl grafts 2 months post-engraftment. a–c UAMPs showing separations of nuclear profiles comparing human versus mouse (a), human neurons versus human astrocytes (b), as well as human APPV717I neurons versus human Ctrl neurons (c). d Heatmap showing the top 30 most variable up- and downregulated DEGs in grafted APPV717I versus Ctrl neurons. e Pathway enrichment analysis of DEGs in grafted APPV717I versus Ctrl neurons. The x-axis shows the normalized enrichment values. The adjusted p value and number of genes are also denoted by color and by size, respectively. f Venn diagram showing up- and downregulated DEGs in APPV717I versus Ctrl neurons in grafts versus in vitro. g Pathway enrichment analysis of DEGs uniquely found in grafted APPV717I neurons. The x-axis shows the normalized enrichment values. The adjusted p value and number of genes are also denoted by color and by size, respectively.
|
This study was performed in collaboration with Drs. Andrew Sproul, Abid Hussaini, Osama Al-Dalahmah and Phil De Jager from the Taub Institute and with Drs. Peter Canoll and Markus Siegelin from the Department of Pathology and Cell Biology at CUIMC.
Gunnar Hargus, MD, PhD
Assistant Professor of Pathology and Cell Biology
gh2374@cumc.columbia.edu
Vilas Menon, PhD
Assistant Professor of Neurological Sciences (in Neurology and the Taub Institute)
vm2545@cumc.columbia.edu
 |  | |
| Nicholas Ray, PhD | Christiane Reitz, MD, PhD |
Genome-wide association studies (GWAS) have identified over 80 Alzheimer’s Disease (AD) risk loci. Most of these studies have been conducted in non-Hispanic white individuals of European ancestry however, while individuals of African ancestry, who have twice the risk of developing AD, have been vastly underrepresented in research. The Alzheimer’s Disease Genetics Consortium has previously identified 16 ancestry-specific genetic variants in African Americans in studies published by our Taub Institute team (2013) and Kunkle et al. (2021).
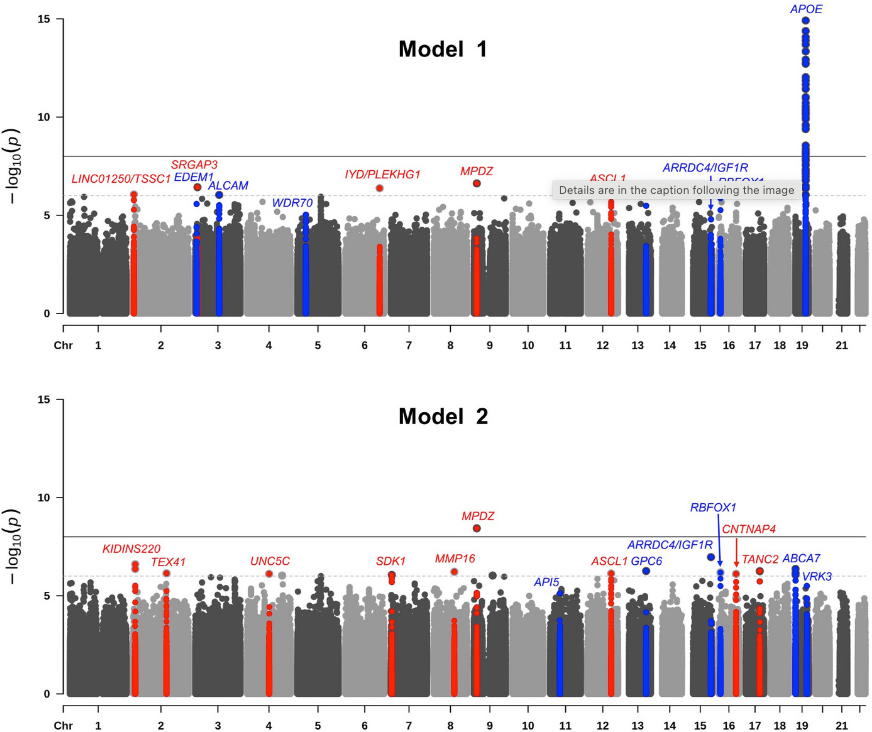
With the addition of three new datasets, including one from Ibadan, Nigeria, the largest GWAS examining AD in individuals of African ancestry increased its sample size by 14.5% to include 9,168 subjects (2,903 AD cases and 6,265 age-matched controls). In the current study, now published in Alzheimer’s & Dementia, our team, including postdoctoral fellow and first author Dr. Nicholas Ray and colleagues from Taub and Neurology, identified a novel risk locus on chromosome 9p23 (rs141610415, P = 3.68×10−9) in the MPDZ gene and 11 additional loci with suggestive associations (P < 9×10−7). MPDZ is highly expressed in the brain and encodes a modular scaffold protein that is localized near the junctions of neuronal synapses. It is involved in regulation of synaptic transmission and is a vital component of the NMDAR signaling complex in excitatory synapses of hippocampal neurons critical for learning and memory.
Pathway analyses identified immunity, lipid processing, intracellular trafficking, DNA repair, and transcription, all of which overlap with pathways shown in non-Hispanic whites. One novel pathway, sodium-independent organic anion transmembrane transport (OAT), did emerge, however. OATs are transporter proteins delivering a range of hydrophobic organic anions including neurotransmitter and amyloid beta metabolites across the blood-brain barrier (BBB). There is significant evidence that BBB dysfunction and dysregulation of BBB transporters are involved in AD etiology.
Our study uniquely included a cohort from continental Africa, which allowed us to compare associations and linkage disequilibrium patterns between datasets with higher and lower degrees of African ancestry. We found differential association patterns in one locus at chromosome 12q23.2 near the ASCL1 gene, suggesting that local ancestry may modulate this association. These findings further illustrate the importance of including populations with diverse genetic ancestry in studies of Alzheimer’s Disease.
Christiane Reitz, MD, PhD
Associate Professor of Neurology and Epidemiology (in the Taub Institute and the Gertrude H. Sergievsky Center)
cr2101@cumc.columbia.edu
 |  |  | ||
| Hilary Grosso Jasutkar, MD, PhD | Ottavio Arancio, MD, PhD | Ai Yamamoto, PhD |
Dysfunction in the lysosome-mediated degradation pathway autophagy has been implicated in various neurodegenerative diseases, including Alzheimer’s disease (AD), yet its specific role in neurological changes remains poorly understood. The question that persists is how can a pathway essential for the function of all organs lead to neurodegeneration above all?
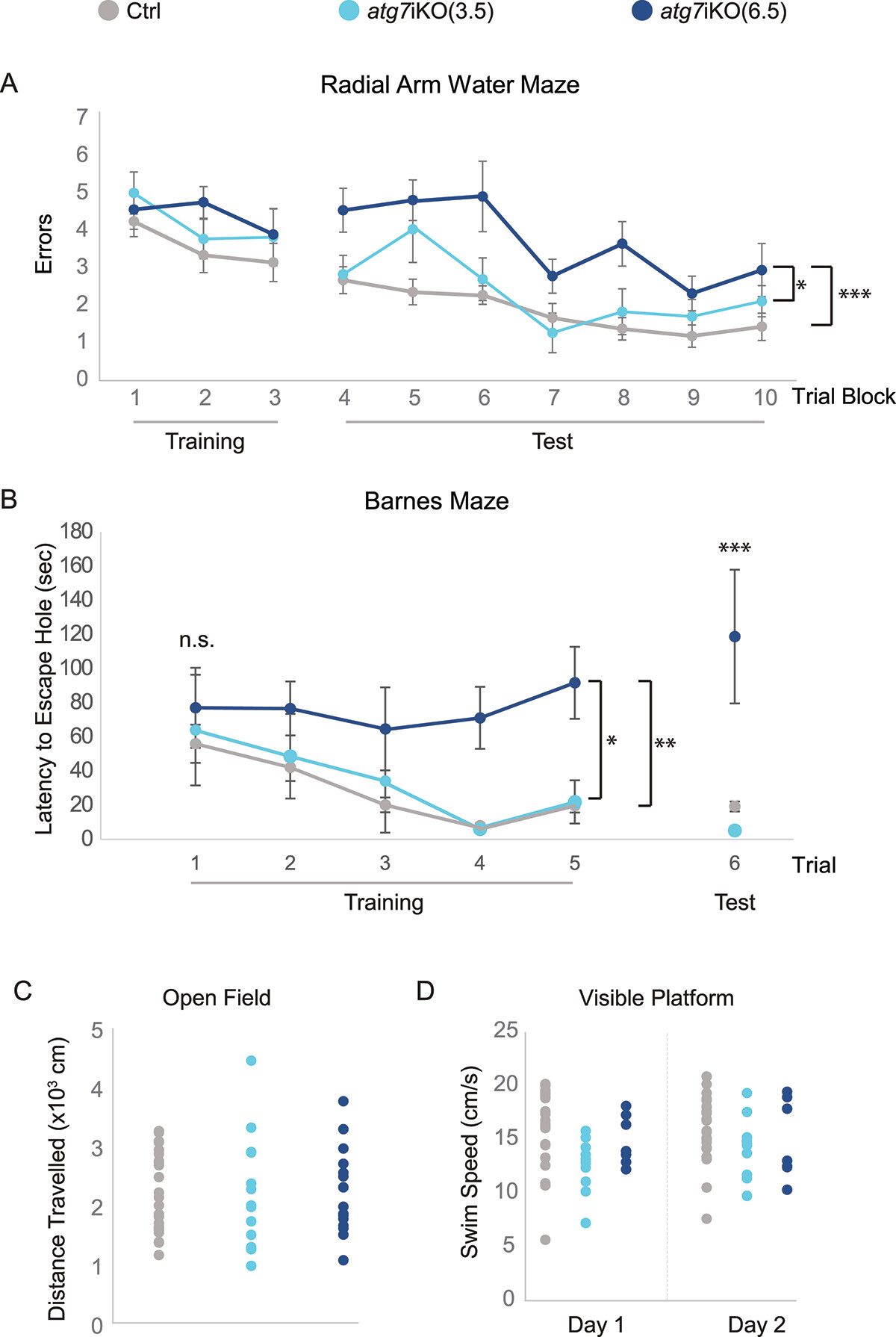
Figure 6. Deactivation of autophagy in adult mice leads to impairment in memory-dependent tasks. (A) 2dRAWM. Over Trial block 1–3 (Training), mice are trained to find a visible platform located in one arm of a 6-arm pool. Mice then must find a hidden platform over Trial blocks 4–5 on day 1, and over Trial blocks 6–10 on day 2. Day 2 is considered the testing day (Test). Although all groups performed similarly during training, only atg7iKO(6.5) mice consistently made more errors than their littermate Ctrls and atg7iKO(3.5) mice during testing. Ctrl, n = 24; atg7iKO(3.5), n = 13; atg7iKO(6.5), n = 10. Mean±St. Dev shown. (B) Barnes maze. Mice are trained to find an escape hole on an open platform over 3 days (Trials 1–5). After a 48-h intertrial interval, they are tested to find the escape hole (Trial 6). On Trials 1–3, all groups exhibited a similar latency to reach the escape hole. With continued Training and on the Test trial, Ctrl and atg7iKO(3.5) were able to acquire and execute the task, whereas atg7iKO(6.5) mice could not acquire the task. Ctrl, n = 16; atg7iKO(3.5), n = 4; atg7iKO(6.5), n = 4. Mean±St. Dev shown. (C and D) Cognitive differences could not be explained by a difference in motor performance. (C) Open Field Maze. Total distance traveled over a 60 min trial. Ctrl, n = 24; atg7iKO(3.5), n = 13; atg7iKO(6.5), n = 10. Individual data points shown. (D) Visible platform test. Daily swim speed average over 6 trials on Day1 and Day 2. Ctrl, n = 23; atg7iKO(3.5), n = 12; atg7iKO(6.5), n = 6. A significant difference in swim speed was found on Day 1 that resolved by Day 2 in the atg7iKO(3.5) group. Statistical analyses are found in Table S1.*p <0.05, **p < 0.01, ***p < 0.001.
To investigate this, our laboratory globally deactivated autophagy in adult mice using the atg7iKO model, with AD-related changes in autophagic processing of synaptic proteins, and then monitored the effects over time. Led by former Columbia Neurology resident and research fellow Dr. Hilary Grosso Jasutkar (now at Rutgers Robert Wood Johnson), we found that synaptosomes isolated from atg7iKO mice showed an accumulation of synaptic proteins, indicating that synapses may be particularly susceptible to disruptions in protein homeostasis. As recently reported in Autophagy, despite the deactivation of autophagy for 6.5 weeks, cognitive impairments were observed without cell death or synapse loss, while motor skills remained unaffected.
Additionally, in collaboration with the Taub Institute laboratory of Dr. Ottavio Arancio, we found that in the APP PSEN1 double-transgenic mouse model of AD, impaired autophagosome maturation and reduced presence of specific synaptic proteins in autophagosomes were noted, leading to the accumulation of the protein SLC17A7/Vglut in the detergent-insoluble fraction. This protein also accumulated in atg7iKO mouse synaptosomes. The findings suggest that synaptic autophagy is important for maintaining protein balance and that its disruption can affect cognitive functions while sparing locomotor abilities, which may be relevant to the cognitive decline seen in AD.
Ai Yamamoto, PhD
Associate Professor of Neurology and Pathology and Cell Biology
ay46@cumc.columbia.edu