Columbia University
Irving Medical Center
Neurological Institute
710 West 168th Street, 3rd floor
(212) 305-1818
TaubCONNECT Research Perspectives:
Best Poster Presentations
Taub Institute Retreat November 2019
Studying Biochemical Mechanisms of Protective Gene Variants in Isogenic Stem Cell-derived Early Onset Alzheimer’s Disease Models
R. Patel, S. Miller, A. Ashok, J. Lee, A. Sproul

Ronak Patel, PhD
The prevailing theory to explain the etiology of Alzheimer’s disease (AD) is the amyloid cascade hypothesis, although attempts to derive disease-modifying therapies that target canonical Abeta production pathways or Abeta itself have failed thus far. The rise in the number of AD patients as people live longer warrants the need for alternative approaches to modify disease progression. Significant progress has been made to identify genes related to AD pathogenesis to understand the mechanism of degeneration. Identification of protective/risk variants and their underlying mechanism is equally important to elucidate novel therapeutic targets against AD. A previous study by Drs. Lee, Mayeux and other colleagues have identified protective variants from genetic screening of a cohort of Puerto Rican families carrying the PSEN1 G206A mutation that is sufficient to cause early onset AD. Single Nucleotide Polymorphisms (SNPs) in SNX25, PDLIM3, SORBS2, SH3RF3 and NPHP1 genes were significantly correlated to age at onset of AD (Lee et. al, JAMA Neurol., 2015). Our goal is to uncover biochemical mechanisms of protection by the aforementioned gene variants in a PSEN1 G206A background by using human neurons derived from isogenic PSEN1 mutant and control pluripotent stem cells (PSCs).
Approach: We have knocked in the PSEN1 G206A mutation into two control PSC lines (H9 and IMR90) using CRISPR-Cas9. Neurons differentiated from PSEN1 G206A and isogenic control PSCs are being used for gain-of-function (overexpression, CRISPRa) and loss-of-function (siRNA) experiments for protective gene variants in neurons derived by Ngn2-mediated transdifferentiation. Amyloid precursor protein (APP) processing and tau levels will be assessed and RNAseq and other “OMIC” assays will be performed to further understand the pathways involved in mechanism of protective gene variants.
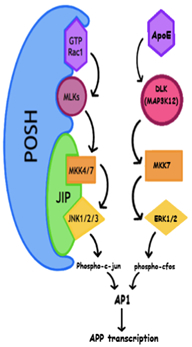
Preliminary Results: Our initial efforts have focused on understanding the role of a protective SNP rs6542814 in SH3RF3/POSH2, which can delay AD onset by 9.3 years in PSEN1 G206A carriers. SH3RF3/POSH2’s paralog POSH has been established as a pro-apoptotic JNK pathway scaffold that facilitates activation of AP-1 (Xu et. al, EMBO 2003). We predicted that a protective SNP in SH3RF3 would reduce its expression. In agreement with this hypothesis, in silico analysis showed that rs6542814 disrupts a binding site for the general transcription factor TFII-I. Although JNK signaling affects many AD-related cellular processes, we became particularly interested assessing effects on APP transcription in light of recent work demonstrating exogenous APOE4 drives APP transcription through AP-1 and non-canonical ERK signaling (Huang et al., Cell 2017). Overexpression of SH3RF3/POSH2 in HEK293T cells drives JNK activation and leads to increased APP transcription. Conversely, siRNA mediated knockdown of SH3RF3/POSH2 in stem cell derived neurons reduces APP transcription. The potential importance of regulation of APP transcription by genetic modifiers of AD is underscored by rare APP triplication that is sufficient to cause early onset AD as well as the high prevalence of dementia in individuals with Down syndrome.
Ronak Patel, PhD
Postdoctoral Research Scientist in the Taub Institute
rp2956@cumc.columbia.edu
Glial Heterogeneity in Normal Human Cortex and Huntington Disease Interrogated Through Single Cell Nuclear RNA Sequencing
 |
| Osama Al Dalahmah, MD, DPhil |
 |
O. Al Dalahmah, A. Sosunov, I. Adorjan, J. E. Goldman
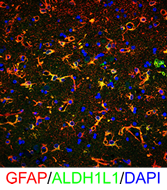
The heterogeneity of CNS glia in the human CNS in normal and pathological conditions requires much further study. To address human glial heterogeneity, we examined transcriptional profiles of control (patients without neurological disease or neuropathology) and advanced Huntington Disease (HD) cingulate cortices using single cell nuclear RNAseq (scnRNAseq) with nuclei isolated from frozen control and HD autopsy tissues. Unsupervised and supervised cluster analysis and classification showed a number of discrete cell clusters corresponding to astrocytes, neurons, oligodendrocytes (OL), OPCs, microglia and endothelial cells. For all cell types, control and HD populations were heterogeneous, and also segregated preferentially between HD and controls. We found a novel signature for reactive HD astrocytes, including a downregulation of NOTCH signaling and fatty acid synthesis genes, and an upregulation of MHC class I and metallothionein (MT) genes. We found 3 different reactive astrocyte “states”, defined by differences in expression of SLC1A2, MTs and GFAP. Astrocytes did not show up-regulation of C3 transcript or C3 protein assessed by immunostaining, but in contrast, astrocytes of the neostriatum of the same brains did upregulate C3 protein, suggesting that different reactions occur in different stages of degenerative pathology in HD. Among OLs we found a large population of OLs in HD that showed marked down-regulation of myelin genes. The HD microglia up-regulated SPP1 (Osteopontin, matrix formation and promotion of inflammation) and HAMP (Hepcidin, iron accumulation), among other genes. All cells upregulated a variety of heat shock protein genes. scnRNAseq provides a technology for interrogating cell-type specific gene expression in the control and pathological CNS from banked brain samples, and the results may identify subset-specific therapeutic targets.
Osama Al Dalahmah, MD, DPhil
Postdoctoral Neuropathology Fellow in the Department of Pathology and Cell Biology
oa2298@cumc.columbia.edu