Columbia University
Irving Medical Center
Neurological Institute
710 West 168th Street, 3rd floor
(212) 305-1818
TaubCONNECT Research Perspectives:
December 2021
2: Atlas of RNA Editing Events Affecting Protein Expression in Aged and Alzheimer's Disease Human Brain Tissue
3: Integration of GWAS and Brain Transcriptomic Analyses in a Multiethnic Sample of 35,245 Older Adults Identifies DCDC2 Gene as Predictor of Episodic Memory Maintenance

Jennifer J. Manly, PhD
The diagnostic category of mild cognitive impairment (MCI) was created to represent a prodromal phase of dementia, but not everyone who is classified as having MCI subsequently progresses to dementia. Longitudinal studies by Taub investigators and others have shown that 5% to 53% of people identified as having MCI at their first visit no longer meet MCI criteria at the next. Therefore, identifying risk factors of progression to dementia in individuals diagnosed with incident MCI is equally important to refine the selection of individuals at high risk for dementia.
The current study, recently published in Neurology, extends our previous findings by investigating modifiable and non-modifiable risk factors of incident MCI among non-Hispanic White, non-Hispanic Black, and Hispanic cognitively normal adults. Together with visiting scholar Dr. Milou Angevaare (Vrije Universiteit Amsterdam), we also examined which factors predict progression to dementia or classification as cognitively normal at follow-up in those who developed incident MCI. Among 2,903 cognitively normal participants at baseline, 752 developed MCI over an average of 6.3 (SD 4.5) years (incidence rate 56 per 1,000 person-years). Presence of APOE ε4 and higher medical burden increased risk of incident MCI, while more years of education, taking part in more leisure activities, and higher income decreased this risk. Of the incident MCI cases, after an average of 2.4 years of follow-up, 12.9% progressed to dementia, 9.6% declined in functioning and did not meet the algorithmic criteria for MCI but did not meet the clinical criteria for dementia, 29.6% continued to meet MCI criteria, and 47.9% no longer met MCI criteria. Multi-domain MCI, presence of APOE ε4, depressive symptoms, and antidepressant use increased the risk of progression to dementia.
Overall, this community-based study showed that almost half of the individuals with incident MCI diagnoses were classified as cognitively normal at follow-up, suggesting the need to refine expectations for cognitive and functional course of those presenting with MCI.
Jennifer J. Manly, PhD
Professor of Neuropsychology (in Neurology, the Gertrude H. Sergievsky Center, and the Taub Institute for Research on Alzheimer’s Disease and the Aging Brain)
jjm71@cumc.columbia.edu
 |  | |
| Yiyi Ma, MD, PhD | Philip L. De Jager, MD, PhD |
RNA editing is a feature of RNA maturation resulting in the formation of transcripts whose sequence differs from the genome template. Brain RNA editing may be altered in Alzheimer’s disease (AD), but there has not yet been a full genome-scale study to systematically analyze gene editing events in relation to AD, or to assess which pathologic processes (amyloid or Tau) or endophenotypes of AD may be associated with RNA editing perturbations.
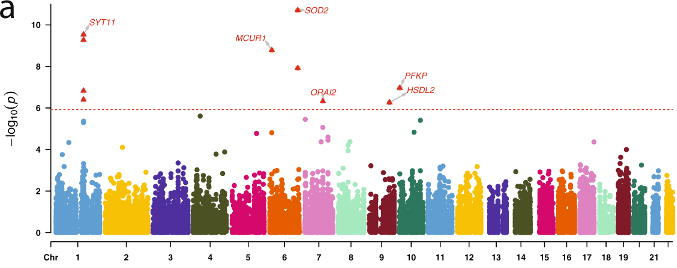
Figure 5. Associations of the top RNA editing events with AD risk and pathologies. (a) Manhattan plot of the Stage II association analysis of RNA editing with clinical status of AD. Each dot represents one RNA editing event, and the X and Y axes show its genomic coordinate and −log10 transformed meta-analyzed P value. The horizontal red dashed line shows the Bonferroni-corrected genome-wide significance threshold (P ≤ 1.2 × 10−6) and those passing the threshold were shown as the red triangles with gene names.
In the current study, recently published in Nature Communications, we analyzed data from 1,865 brain samples covering 9 brain regions from 1,074 unrelated subjects on a transcriptome-wide scale to identify inter-regional differences in RNA editing. We expand the list of known brain editing events by identifying 58,761 previously unreported events. Of note, only a small proportion of these editing events were found at the protein level in their proteome-wide validation effort. We also identified the occurrence of editing events associated with AD dementia, neuropathological measures and longitudinal cognitive decline in: SYT11, MCUR1, SOD2, ORAI2, HSDL2, PFKP, and GPRC5B.
Overall, we present an extended reference set of brain RNA editing events, identify a subset that are found to be expressed at the protein level, and extend the narrative of transcriptomic perturbation in AD to RNA editing. This work builds upon our team’s recent report of reproducible splicing alterations in AD, and the broader narrative of specific disruptions in the epigenome in relation to AD.
Yiyi Ma, MD, PhD
Instructor in Neurological Sciences (in Neurology) at CUMC
Center for Translational & Computational Neuroimmunology
Taub Institute for Research on Alzheimer’s Disease and the Aging Brain
ym2666@cumc.columbia.edu
Philip L. De Jager, MD, PhD
Weil-Granat Professor of Neurology (in Neurology, the Taub Institute for Research on Alzheimer's Disease and the Aging Brain, and the Precision Medicine Initiative)
pld2115@cumc.columbia.edu
 |  | |
| Yizhe Gao, MS | Sandra Barral, PhD |
Major research studies at Columbia and elsewhere have shown that the effects of aging on memory performance are very heterogeneous, with clear inter-individual vulnerabilities. Given the healthcare crisis posed by our aging world population, it is increasingly important to understand the causal factors underlying changes in episodic memory performance as individuals age. In the current study, we integrated common and rare genetic variants and transcriptomics data in an effort to identify novel episodic memory loci that might help distinguish the memory decline attributable to normal aging from that indicating pathological aging.
Building on previous work, episodic memory trajectories estimated in an ethnically diverse sample of 35,349 elderly were used as outcome to conduct apolipoprotein E (APOE)-stratified genome-wide association study (GWAS) analyses. As recently reported in Alzheimer’s & Dementia, data integration of GWAS meta-analysis and brain expression results provided evidence for the DCDC2 gene as a novel predictor of memory maintenance among non-carriers of APOE-ε4 (N=24,941). Further, brain transcriptomics revealed an association between episodic memory maintenance and (1) increased dorsolateral prefrontal cortex DCDC2 expression and (2) lower burden of pathological Alzheimer's disease (AD) hallmarks. Moreover, when AD cases were compared to cognitively healthy participants, DCDC2 expression was decreased across all brain areas.
The DCDC2 gene was previously reported as significantly associated with general cognitive function in a sample of more than 300,000 subjects from European cohorts including UKBB. The DCDC2 gene is one of the most conserved genes of the doublecortin superfamily, a group of proteins that regulate filamentous actin structure in developing neurons. DCDC2 binds to tubulin and enhances microtubule polymerization influencing synaptic plasticity. The identification of the DCDC2 gene as a predictor of memory maintenance in older adulthood provides the possibility of identifying population subgroups at-risk of memory decline and dementia, paving the way for precision medicine intervention.
Yizhe Gao, MS
Former Staff Research Associate
Columbia University Irving Medical Center
yzhegao@gmail.com
Sandra Barral, PhD
Associate Professor of Neurogenetics (in Neurology, the Gertrude H. Sergievsky Center, and the Taub Institute for Research on Alzheimer’s Disease and the Aging Brain) at CUMC
smb2174@cumc.columbia.edu