Columbia University
Irving Medical Center
Neurological Institute
710 West 168th Street, 3rd floor
(212) 305-1818
Featured Research
In the Lab:
Estela Area Gomez, PhD

Estela Area Gomez, PhD
My laboratory focuses on the study of lipid metabolism in the nervous system, and the role of alterations in lipid homeostasis in the pathogenesis of neurodegenerative disorders, such as Alzheimer's disease, traumatic brain injury, and amyotrophic lateral sclerosis.
Specifically, our data shows that C99, a product of amyloid-╬▓ precursor protein (APP) processing, is involved in the regulation of cholesterol trafficking and sphingolipid turnover. To understand the mechanism behind this role of C99, we are using a combination of unbiased and targeted proteomics analyses to identify the enzymes involved in the pathways of cholesterol and sphingolipid metabolism that are regulated by mitochondria-associated membranes (MAM)-C99. Moreover, we are assessing the role of C99 in the binding of cholesterol-containing endosomes to the endoplasmic reticulum (ER) by imaging and trafficking studies in our cell models.
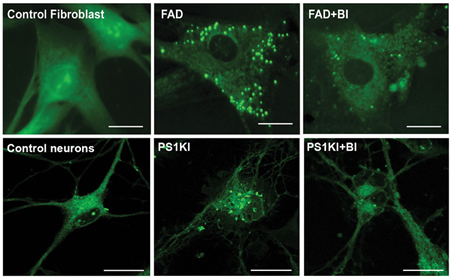
Lipid droplets in AD cells. Staining of lipid droplets in AD cells (PS1A246E) and cortical neurons from WT and PS1-KIM146V mice. Note reduction in LDs after inhibiting BACE1 (BI). Size bars=20╬╝m. Quantification on the right (n=6 biological replicates. 500 cells /replicate ┬▒S.D. * p<0.05 T-test)
By the same token, our working hypothesis maintains that increased levels of C99 are at least partly responsible for the lipid abnormalities observed in Alzheimer's disease (AD). To examine this, our work within the Taub Institute is focused on using AD cell models and tissues to measure levels of C99, and analyzing the changes in the lipid composition of cellular membranes from these models. Our goal is to understand and explain the role of C99 in AD, and the effect of changes in the membrane lipid composition in propagating known AD phenotypes.

Our team and collaborators: (from left, top row) Estela Area Gomez (PI), Kristin Tamucci, Delfina Larrea, Fereshteh Zandkarimi (Stockwell Lab), Cristina Guardia-Laguarta (Przedborski Lab), Kevin Velasco; (from left, bottom row) Taekyung "Daniel" Yun, Rishi Agrawal, Yimeng "Oliver" Xu (Lipidomics Facility), and Jorge Montesinos.
Finally, we have developed a new algorithm to analyze data from untargeted lipidomics analysis to unveil a "snap-shot" of the metabolic status of a cell or tissue, and unravel alterations in specific cellular processes. Our preliminary results show specific signatures in cells and serum samples from AD that point toward specific alterations in the pathway that govern cholesterol trafficking and the regulation of phospholipid acylation. We aim to perform longitudinal lipidomics analysis to understand the timeline of these lipid alterations, toward the development of a new diagnostic methodology based on more than 600 lipid species. Our goal, with this work, is to unravel new therapeutic targets, toward future therapies that might prevent dementia in the earliest stages.