Columbia University
Irving Medical Center
Neurological Institute
710 West 168th Street, 3rd floor
(212) 305-1818
Featured Research
In the Lab:
Luke E. Berchowitz, PhD

Luke E. Berchowitz, PhD
Amyloids are fibrous protein assemblies that are associated with numerous human diseases including Alzheimer's disease. Although amyloids have been predominantly understood in pathological contexts as toxic and irreversible protein deposits, amyloids can have critical physiological functions (Fowler et al., 2007). The goals of my lab are to determine the processes that healthy cells have developed to regulate the accumulation and/or reversibility of amyloids. We are also interested in understanding the molecular mechanisms underlying how these structures function in biological processes. Elucidating these mechanisms is important to advance our understanding of the diseases associated with amyloids and, in time, could lead to therapeutic opportunities.
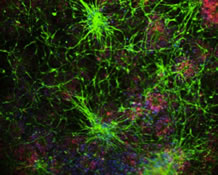
My lab primarily uses yeast cells as model system to investigate the regulation and function of amyloids. How can yeast inform the biology of the aging brain? Yeast cells have the remarkable ability to build assemblies (of a protein called Rim4) that are structurally similar to the amyloids that form in the brains of individuals suffering from Alzheimer’s disease (Figure 1) (Berchowitz et al., 2015). Due to the similarity of these structures to disease-related amyloids they are termed 'amyloid-like.'
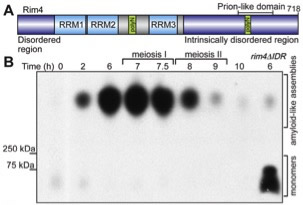
Figure 1. Rim4 forms amyloid-like assemblies during meiosis. (A) Diagram of Rim4 (B) Meiotic time course analysis of Rim4 amyloid-like assemblies. A hallmark property of amyloids is resistance to strong detergents such as SDS. Samples were incubated in 2% SDS for 10 min and resolved on 1.7% agarose (with 0.1 SDS). Rim4 was detected by immunoblot analysis. Rim4 amyloid-like assemblies migrate as large particles that are cleared after meiosis I. Rim4 lacking its C-Terminal IDR is shown as a control.
What is remarkable about amyloid-like assemblies in yeast is that they are not pathogenic, on the contrary, they are functional, regulated, and essential for sexual reproduction. Beyond yeast, transient amyloid-like assemblies are also critical features of the reproductive cycle in mouse and frog highlighting the evolutionary conservation of these structures (Berchowitz et al., 2015; Boke et al., 2016). In my lab, we have begun to understand the mechanisms underlying yeast’s ability to disassemble and clear these structures (Carpenter et al., 2018) (Figure 2). Our results have opened the door for a new understanding of how healthy cells can protect against or even reverse pathological amyloids.
Moving forward, our lab will continue to use genetic, biochemical, and cell biological approaches to identify and characterize the genes and pathways that cells use to regulate amyloid and amyloid-like assemblies. We are also working to elucidate how environmental factors influence amyloid accumulation.
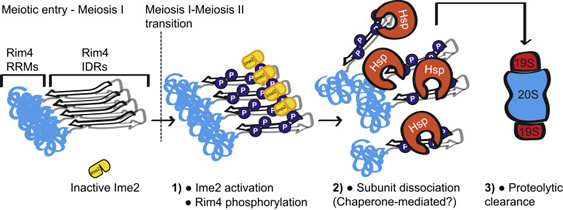 Figure 2. Model for phosphorylation-driven reversibility and clearance of amyloid-like assemblies in meiosis
|
In order to expand our knowledge from yeast to human disease, we need a model system in which a disease-related amyloid is reversed. For this purpose, we are developing a new model system: ground squirrel cells. Ground squirrels accumulate tau amyloid fibers (a hallmark of Alzheimer's disease) in their brain cells during hibernation (Arendt et al., 2003). These structures are reversed upon exit from hibernation. Stem cells derived from ground squirrel can recapitulate much of the biology underlying the hibernating state (Ou et al., 2018). By studying amyloid reversibility in these cells, we can unlock a deep understanding of the genes, proteins, and mechanisms that are required to reverse amyloids.
 From left to right, members of the Berchowitz lab include Grace Herod,
Kayla Carpenter,
Diana Ottoz, PhD,
Luke Berchowitz, PhD,
Julius Yunus, and
Raphaelle Laureau, PhD.
|
Luke E. Berchowitz, PhD
Assistant Professor of Genetics and Development
leb2210@cumc.columbia.edu