Columbia University
Irving Medical Center
Neurological Institute
710 West 168th Street, 3rd floor
(212) 305-1818
Featured Research
In the Lab:
Falak Sher, PhD

Falak Sher, PhD
Genetic studies have discovered many genomic loci/variants that are associated with Alzheimer’s disease (AD). However, biological interpretation of these associations is generally not straightforward. Our aim is to perform detailed dissection of the effect of AD associated variants/loci on cellular function and phenotype using state of the art functional genomics tools. One approach is that we are pursuing entails the functional annotation of AD-associated genomic risk genes/loci by using next generation CRISPR tools, e.g. CRISPRa and CRISPRi.
A complementary interest is to identify the rational drug targets within multimeric protein complexes that contain AD-associated protein(s). To accomplish these goals, we are integrating next generation CRISPR/Cas9 genome editing tools, targeted quantitative proteomics, and high dimensional data sets from aging human brains. We use human dorsolateral prefrontal cortex tissues and purified immune/glial cells from autopsy brain material (ROS-MAP/New York Brain Bank), as well as in vitro model systems.
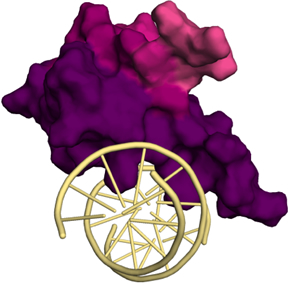
Figure 1. PDB 6CNQ is colored based on functional scores obtained from CRISPR mediated functional dissection of MBD2, a NuRD subunit. The amino acids that are in physical proximity of the DNA are more effective (darker color) in terms of function (i.e. gene repression).
During my postdoctoral training in the laboratory of Dr. Daniel E. Bauer at Harvard Medical School, we developed a generalizable CRISPR-based technology for comprehensive functional dissection of multi protein complexes for parallel ascertainment of structure-function relationships or to uncover the gene expression regulatory architecture of a target protein. More broadly, this approach produces functional maps of co-complexed proteins and regulatory regions (e.g. promoters, enhancers) virtually at amino acid and nucleotide resolution, respectively. These maps pinpoint the amino acids or nucleotides responsible for critical protein-protein and protein-DNA physical interactions (Figure 1). This information enables rational chemical biology experiments to investigate small molecules that interfere with identified critical interactions, which will pave the way for targeted drug development. Currently, in collaboration with Dr. Richard Mayeux and Dr. Philip L. De Jager, we are using this method to investigate the mechanistic and structural role of SORL1, PLCG2 and ABI3 in health and disease. These AD risk genes are predominantly expressed in microglia, the resident immune cells of the brain. We have identified many novel physical interactions of these proteins which could be critical in AD pathogenesis. This pipeline, comprised of CRISPR mediated dissection of disease associated loci, will be subsequently used to study all AD-associated GWAS hits in a similar fashion.
Another parallel line of research that we are perusing is the construction of a chromatin map of aging microglia and the systematic functional dissection of microglial chromatin structure by integrating genome editing and ATAC-Seq methodologies. Epigenome studies of brain tissue have identified enrichment of AD-associated loci in enhancer regions, and chromatin accessibility studies (ATAC-Seq) have demonstrated microglia specific active enhancers containing AD-associated polymorphism (e.g. BIN1). Thus, detailed understanding of the architecture of microglial chromatin could help us to prioritize the investigation of known and suggestive AD loci. To get further insight into the epigenetic changes which can affect the microglia phenotypes in physiological conditions and in AD pathogenesis, we are modulating epigenetic readers and writers by using various modified CRISPR/Cas9 systems.
Overall, these efforts will help to unlock the therapeutic potential of AD associated genetic discoveries.
Falak Sher, PhD
Instructor in Neurological Sciences (in Neurology and the Taub Institute for Research on Alzheimer's disease and the Aging Brain) at CUIMC
fs2644@cumc.columbia.edu