Columbia University
Irving Medical Center
Neurological Institute
710 West 168th Street, 3rd floor
(212) 305-1818
Featured Research
In the Lab:
Radhika Jagannathan, MD, PhD

Radhika Jagannathan, MD, PhD
Neurodegenerative diseases such as Parkinson’s disease (PD) and Alzheimer’s disease (AD) are among the most common aging-related disorders, imposing significant personal, social, and monetary costs on patients and their families. Gene expression analysis from autopsy specimens has been a valuable tool to identify cellular pathways that are dysregulated in and may contribute to neurodegenerative disease. However, the pathogenic upstream mechanisms leading to these gene expression changes remain largely unknown. Transcription factors (TFs) are important cellular components that govern gene expression through protein-DNA interactions. Several TFs can form dimers or multimeric complexes and the transcriptional effect of a given TF can be modulated by its choice of binding partner. This combinatorial regulation of TFs allows them to participate in diverse cellular processes in different contexts and may help explain the clinical heterogeneity and cell-type specificity associated with different neurodegenerative disorders. As a cell biologist and neurologist by training, I am interested in understanding how disease-specific TF complexes contribute to neurodegenerative pathology and cellular dysfunction.
I came to Columbia University in 2018 as a Clinical Fellow in Behavioral Neurology. During my fellowship, I have been working in the Taub Institute laboratory of Dr. Ulrich Hengst, studying a specific TF heterodimer consisting of the stress-responsive TF, ATF4, and a newly-identified binding partner, CREB3L2. A colleague in Dr. Hengst’s lab, Dr. Claudio Roque, has identified this TF complex as an important regulator of gene expression in Alzheimer’s disease. However, ATF4 has also been implicated in the pathogenesis of PD; it is upregulated in the substantia nigra of PD patients, as well as in numerous cellular PD models, suggesting that the ATF4-CREB3L2 heterodimer may also be important for gene dysregulation in PD. Consistent with this, I have found that both ATF4 and CREB3L2 are upregulated and that there is increased formation of the ATF4-CREB3L2 complex in the substantia nigra of PD patients. Using a bioinformatics approach with existing gene expression datasets from PD patients, I have been able to identify cellular processes that are likely to be dysregulated by this TF complex in PD. This approach also identified several genes as potential ATF4-CREB3L2 targets, including mitochondrial genes such as CHCHD2, and the ER-to-Golgi vesicular transport regulator RAB1A.
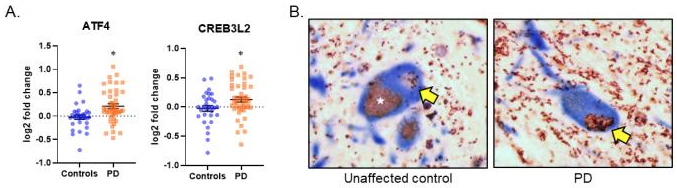 Figure 1. Increased expression and formation of the ATF4-CREB3L2 TF dimer in PD
(A) A meta-analysis of 7 published microarray expression datasets of substantia nigra (SN) from PD and control patients was performed to assess for ATF4 and CREB3L2 expression. (B) Proximity ligation assay for the ATF4-CREB3L2 complex in PD and control SN. Red-brown puncta indicate an interaction event. Nuclei containing puncta are highlighted by the arrow. Tyrosine hydroxylase is stained in blue, with nuclei in purple. Lighter brown areas inside neurons (☆) are neuromelanin. Sections obtained from The New York Brain Bank at Columbia University. * = p < 0.05.
|
I will be completing my fellowship in June 2021 and am excited to be staying on as a clinical instructor in the Department of Neurology, and continuing my research in Dr. Hengst’s lab. Going forward, I plan to validate the importance of ATF4-CREB3L2 on mitochondrial function and intracellular transport using cellular PD models. I also am interested in identifying other PD-specific ATF4 TF dimers to better understand how differential heterodimerization of TFs can modulate cellular function. My hope is that the identification of disease-specific TF dimers might outline an approach to selectively target pathogenic gene expression changes while leaving the regular functions of TFs intact.
Radhika Jagannathan, MD, PhD
Clinical Fellow, Division of Aging and Dementia/Laboratory of Dr. Ulrich Hengst
rj2535@cumc.columbia.edu